Preliminary analysis or your work on the Relationship Extraction task
We have been working on a preliminary analysis of the relationship extraction data generated by our fantastic Mark2Curators. This analysis is in the process of being written up into an academic paper and if your data was used for this paper, you will be given option of being credited on a page on our site dedicated to the contributors for this paper. We expect to have an email notifying contributors to the data set out by early next week, so keep an eye out for it.
San Diego Hackathon / Curation Jamboree
If you are in San Diego the week of October 14th, and have a background in software development, engineering, computational biology, bioinformatics, pathology, oncology, genomics, or biocuration--there is a hackathon/curation jamboree happening on October 15th-October 16th. The event is a joint event between the Griffith Labs, Su and Wu Labs and will cost $25 to register. Mark2Cure is a project of the Su Lab focusing on biomedical literature curation; while, the hackathon is focused on the CIViC resource from the Griffith labs. CIViC is an open access, open source, community-driven web resource for Clinical Interpretation of Variants in Cancer which aims to enable precision medicine by providing an educational forum for dissemination of knowledge and active discussion of the clinical significance of cancer genome alterations. You can learn more about this event at: https://www.eventbrite.com/e/cancer-variant-interpretation-hackathon-and-curation-jamboree-tickets-48287431006?aff=General
World Alzheimer Day
In case you missed it, September 21st is World Alzheimer’s day, and our friends at EyesOnAlz will be holding a world-wide Catchathon. Our very own TAdams organized a local team to participate in a previous Catchathon. If members of the Mark2Cure community are interested in teaming up and contributing with other Mark2Curators for this Catchathon, we would be happy to organize a Mark2Cure team for the event. Otherwise, if you are interested in contributing to Alzheimer’s research from the comfort of your own computer on an individual basis, we 100% encourage you to do so!
Science by the people and for the people—introducing a new Citizen Science project from the Knight Lab at UCSD
If you’ve ever wished that there was a citizen science project for answering questions about how nutrition and other habits affect health and other outcomes—there’s now a new platform to address your questions. This platform, Galileo, comes from the Knight Lab at the University of California, San Diego—the same lab that is responsible for the American Gut project!
Here’s how it works:
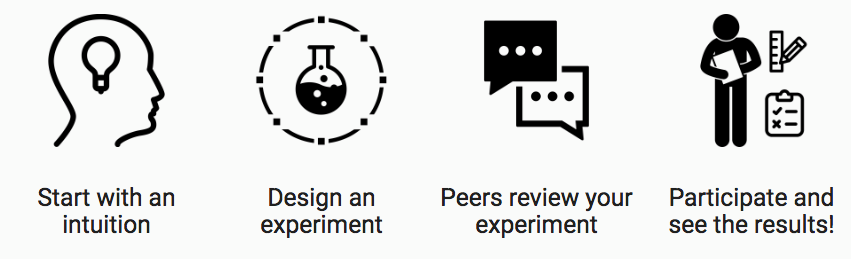 Learn more at gutinstinct.ucsd.edu/info
Learn more at gutinstinct.ucsd.edu/info


 Interestingly enough, Lipman's and the GenBank's team nomination for the 2017 Sammies only cursorily mention PubMed Central in favor of focusing on GenBank and his contributions to infectious disease surveillance. Perhaps describing their work this way made it more accessible to anyone not in biomedical research. Unfortunately,
Interestingly enough, Lipman's and the GenBank's team nomination for the 2017 Sammies only cursorily mention PubMed Central in favor of focusing on GenBank and his contributions to infectious disease surveillance. Perhaps describing their work this way made it more accessible to anyone not in biomedical research. Unfortunately, 

